Using droplet fluidics to mimic hypoxia and nutrient deprivation conditions of tuberculosis; an original bubble act
By LifETIME CDT Student: Antonia Molloy (Aston University)
Tuberculosis (TB) is the world’s leading cause of death by infectious disease. As a young researcher learning immunology, I was particularly fascinated by the ability of Mycobacterium tuberculosis (the causative agent of TB disease) to manipulate the hosts’ own immune system for its survival. Instead of protecting the body, immune cells are manipulated to protect the bacteria in an aggregation of cells known as a granuloma. Here, bacteria can survive in a ‘dormant’ non-replicating persistent (NRP) state in oxygen and nutrient deprived conditions. It is this physiology which makes it challenging to therapeutically target the bacteria with antibiotics. Up to one third of the world’s population is estimated to be latently infected with M. tuberculosis, which can relapse into active TB in high-risk individuals. This interest led me to undertake a multi-disciplinary PhD including the fields of microbiology and engineering, with the objective to tackle the looming crisis of TB.
Poor Experimental Models
Culturing of bacterial cells is important for antimicrobial susceptibility testing when investigating the activity of new antibiotic compounds or combinations. Conventional culturing techniques include laborious workflows using microtiter plates and petri dishes, having output of a few hundred data points. Additionally, traditional culturing methods may not be indicative of in vivo human physiology and therefore does not convey the ideal pharmacokinetic and pharmacodynamic effects of drugs. The current in vitro models for studying the NRP state of TB do not represent fully the physiological state of the cell and tissue environment (Gibson et al., 2018). We desperately require validated high-throughput models that predict human outcomes of drug action against this slow growing pathogen. Engineering technology has the potential to drive this forward.
Droplet Fluidics
Microfluidics is the study of fluids at the micro, nano, or even pico scale. Fluid behaviour, manipulation, and application can easily be studied in multi-step reactions. Microfluidics has been elegantly used in TB research including diagnostics, experimental modelling, and drug discovery. A new emerging field of microfluidics is droplet fluidics. Utilising flow dynamics, droplets (‘bubbles’) can be created on a microfluidic chip. Three components are required to develop these bioreactors: oil, a stabilising biocompatible surfactant, and an experimental sample. These water-in-oil droplets have many advantages which are listed in Figure 1. The applications of droplet fluidics in microbiology research have been extensively reviewed and involve: detection and identification of microorganisms in droplets, antimicrobial susceptibility analysis, and microbial physiology and interactions. This new era of advanced technology allows the study of bacteria at the single cell level.
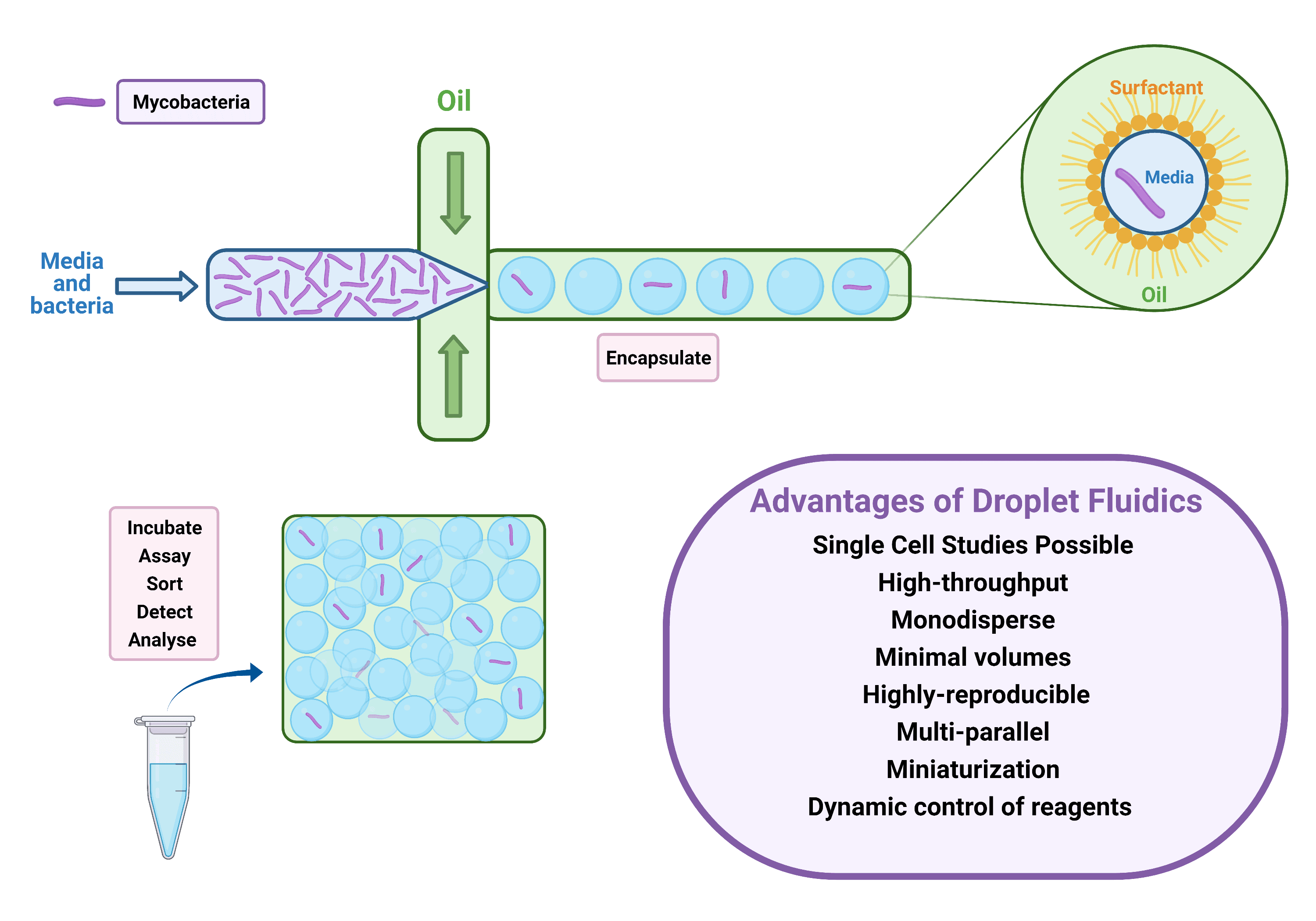
Figure 1: Droplet Generation
Project Aims
I will be undertaking a multi-disciplinary approach combing the technological advancement of microfluidics and microbiology. A new in vitro model mimicking the NRP state of TB will be designed. Using droplet fluidics, an appropriate and safe model organism of M. tuberculosis will be encapsulated at the single cell level. The physiologically relevant growth conditions of the bacteria will be controlled in these droplet bioreactors. Once the in vitro model is optimised, I will take a similar approach as that taken by Liu et al., 2016 and screen new potential therapeutic agents
against this organism as well as establishing their mechanism of action. Scientific questions and investigations include the method of generating the droplets through chip geometry and flow rate. Additionally, the droplet system will be explored comprising encapsulation, incubation, and interrogation of droplets. Droplet bioreactors will be controlled with relevant environmental conditions before the output characteristics (throughput and accuracy) are optimised for the new developed drug screening assay (Figure 2).
This project collaborates with an industrial partner in Cambridge – Sphere Fluidics – who are experts in the droplet fluidic field. Their blog post which can be found here, explains how their ‘picodroplet’ technology is overcoming the current synthetic biology workflow challenges.
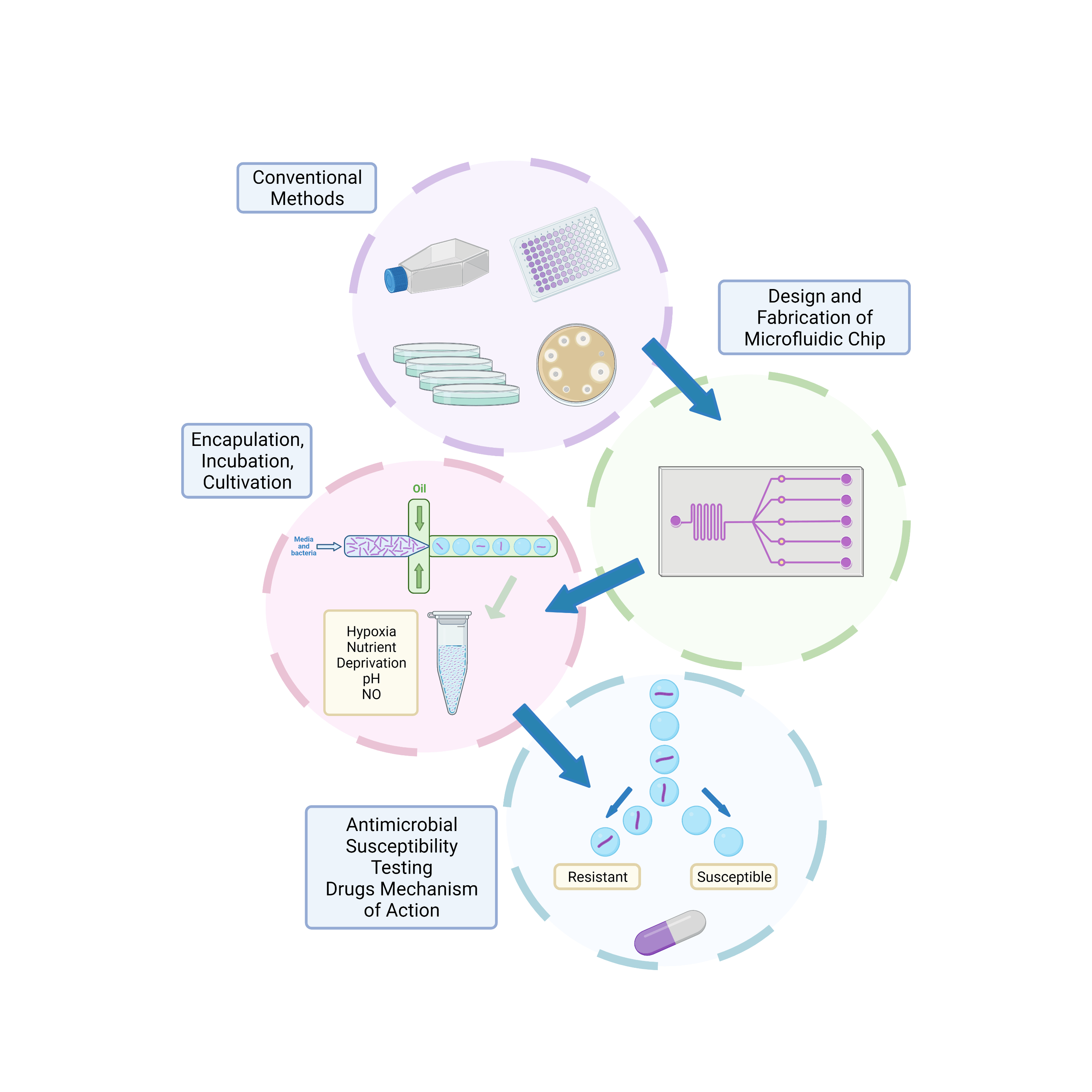
Figure 2: Project Workflow
Impact and Future Applications
What impact could this model provide? Recreating antimicrobial susceptibility testing for NRP TB interests many stakeholders including drug makers and regulatory agencies. High-throughput and physiologically relevant data sets will enable stronger statistical data for regulators to accept new drugs and speed up the antimicrobial discovery pipeline. With all the ‘low hanging fruit’ having been investigated, a shortage of blockbuster drugs has called for high-speed biological assays to push towards feasible clinical trials.
The open research model, with an industrial and academic collaboration, will strengthen the healthcare and technology landscape. In his blog, Charlie House (an Engineering and Physical Sciences Research Council doctoral student) reflected on his experience of doing a joint project with academia and industry. He explains the partnership brought additional opportunities and insight to his work. My project will join forces with Sphere Fluidics to gain mutual benefit in open innovation for engineering and drug discovery.
Future applications of my optimised model may include personalised treatment to patients by using this technology to test susceptible and resistant strains of the bacteria and choose appropriate treatment options. Giving the ‘right drug, to the right patient, at the right time’, affiliates with the new era of precision medicine.
The project aims also align with the interests of this generation – less plastic, reduced chemicals, and the minimalisation of animal models. Our work closely follows the principles of greener, more sustainable and less environmentally damaging research. I am eager to develop this innovative in vitro platform to address the current health threat of tuberculosis.
References
Gibson, S. E. R., Harrison, J., & Cox, J. A. G. (2018). Modelling a Silent Epidemic: A Review of the In Vitro Models of Latent Tuberculosis. Pathogens (Basel, Switzerland), 7(4), 88. https://doi.org/10.3390/pathogens7040088
Liu, X., Painter, R. E., Enesa, K., Holmes, D., Whyte, G., Garlisi, C. G., Monsma, F. J., Rehak, M., Craig, F. F., & Smith, C.A. (2016). High-throughput screening of antibiotic-resistant bacteria in picodroplets [10.1039/C6LC00180G]. Lab on a Chip, 16(9), 1636-1643. https://doi.org/10.1039/C6LC00180G